TRANSPOSITION AND CONSERVATIVE SITE-SPECIFIC RECOMBINATION
We have seen that homologous recombination can result in the exchange of DNA sequences between chromosomes. However, the order of genes on the interacting chromosomes typically remains the same after homologous recombination, inasmuch as the recombining sequences must be very similar for the process to occur. In this part of the chapter, we describe two very different types of recombination—transposition and conservative site-specific recombination—that do not require substantial regions of DNA homology. These two types of recombination reactions can alter the gene order along a chromosome and introduce whole blocks of DNA sequence into the genome.
Transposition and conservative site-specific recombination are largely dedicated to moving a wide variety of specialized segments of DNA—collectively termed mobile genetic elements—from one position in a genome to another. We will see that mobile genetic elements can range in size from a few hundred to tens of thousands of nucleotide pairs, and each typically carries a unique set of genes. Often, one of these genes encodes a specialized enzyme that catalyzes the movement of only that element and its close relatives, thereby making this type of recombination possible.
Virtually all cells contain mobile genetic elements, known informally as “jumping genes.” As explained in Chapter 4, over evolutionary time scales, they have had a profound effect on the shaping of modern genomes. For example, nearly half of the DNA in the human genome can be traced to these elements (see Figure 4–63). Over time, random mutation has altered their nucleotide sequences, and, as a result, only a few of the many copies of these elements in our DNA are still active and capable of movement. The remainder are molecular fossils whose existence provides striking clues to our evolutionary history.
Mobile genetic elements are often considered to be molecular parasites (they are also termed “selfish DNA”) that persist because cells cannot get rid of them; they certainly have come close to overrunning our own genome. However, mobile DNA elements can provide benefits to the cell. For example, the genes they carry are sometimes advantageous to the host, as in the case of antibiotic resistance in bacterial cells, discussed later. The movement of mobile genetic elements also produces many of the genetic variants upon which evolution depends, because, in addition to moving themselves, mobile genetic elements occasionally rearrange neighboring sequences of the host genome. Thus, spontaneous mutations observed in bacteria, Drosophila, humans, and other organisms are often due to the random movement of mobile genetic elements. While many of these mutations will be deleterious to the organism, some will be advantageous and may spread throughout the population. It is almost certain that much of the variety of life we see around us originally arose from the movement of mobile genetic elements.
In this part of the chapter, we introduce mobile genetic elements and describe the mechanisms that enable them to move from place to place in a genome. As mentioned above, these elements move through a variety of different mechanisms that can be grouped into two broad categories, transposition and conservative site-specific recombination. We begin with transposition, by far the most predominant of these two processes.
Through Transposition, Mobile Genetic Elements Can Insert into Any DNA Sequence
Mobile elements that move by way of transposition are called transposons, or transposable elements. In transposition, a specific enzyme, usually encoded by the transposon itself and typically called a transposase, acts on specific DNA sequences at each end of the transposon, causing it to insert into a new DNA site. Most transposons are only modestly selective in choosing their target site, and they can therefore insert themselves into many different locations in a genome; in particular, there is no general requirement for sequence similarity between the ends of the element and the target sequence. Most transposons move only rarely. In bacteria, the rate is typically one transposition event once every 105 cell divisions, and significantly more frequent movement would probably destroy the host cell’s genome. In plants and animals, the situation is different: it is common for progeny to carry tens to hundreds of new insertions relative to their parents. These high rates are tolerated, in part, because these genomes typically carry vast amounts of nonessential DNA sequences where most of the insertions are likely to occur.
On the basis of their structure and transposition mechanism, transposons can be grouped into three large classes: DNA-only transposons, retroviral-like retrotransposons, and nonretroviral retrotransposons. The differences among them are briefly outlined in Table 5–4, and each class will be discussed in turn.
DNA-only Transposons Can Move by a Cut-and-Paste Mechanism
DNA-only transposons, so named because they exist exclusively as DNA during their movement, predominate in bacteria, and they are largely responsible for the spread of antibiotic resistance in bacterial strains. When antibiotics such as penicillin and streptomycin first became widely available in the 1950s, most bacteria that caused human disease were susceptible to them. Now, the situation is different—antibiotics such as penicillin (and its modern derivatives) are no longer effective against many modern bacterial strains, including those causing gonorrhea and bacterial pneumonia. The spread of antibiotic resistance is due largely to genes that encode antibiotic-inactivating enzymes that are carried on transposons (Figure 5–58). Although these mobile elements can transpose only within cells that already carry them, they can be moved from one cell to another through other mechanisms known collectively as horizontal gene transfer (see Figure 1–18). Once introduced into a new cell, a transposon can insert itself into the genome and be faithfully passed on to all progeny cells through the normal processes of DNA replication and cell division.
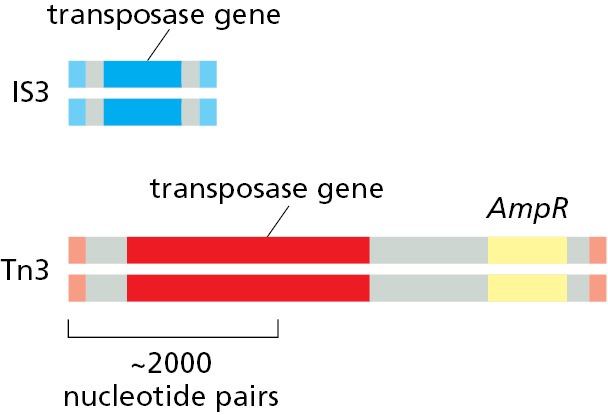
Some transposons carry additional genes (yellow) that encode enzymes that inactivate antibiotics such as ampicillin (AmpR). The spread of these transposons is a serious problem in medicine, as it has allowed many disease-causing bacteria to become resistant to the antibiotics developed in the twentieth century.
DNA-only transposons can relocate from a donor site to a target site by cut-and-paste transposition (Figure 5–59). Here, the transposon is literally excised from one spot on a genome and inserted into another. This reaction produces a short duplication of the target DNA sequence at the insertion site; these direct repeat sequences that flank the transposon serve as convenient records of a prior transposition event. Such “signatures” often provide valuable clues in identifying transposons in genome sequences.
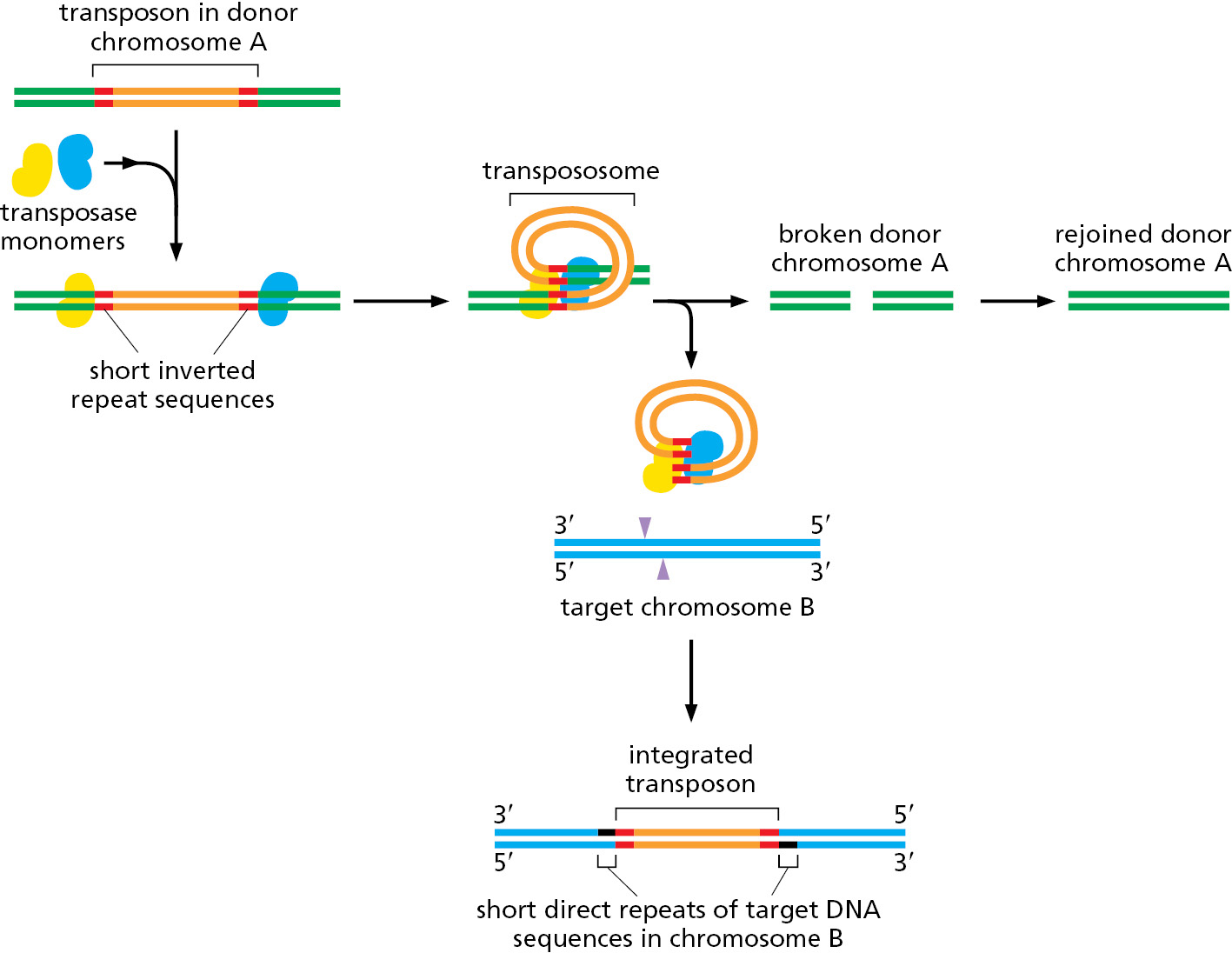
When a cut-and-paste DNA-only transposon is excised from its original location, it leaves behind a “hole” in the chromosome. This lesion can be perfectly healed by recombinational double-strand break repair, provided that the chromosome has recently been replicated so that an identical copy of the damaged host sequence is available. Alternatively, a nonhomologous end-joining reaction can reseal the break; in this case, the DNA sequence that originally flanked the transposon is often altered, producing a mutation at the chromosomal site from which the transposon was excised (see Figure 5–45).
Remarkably, the same mechanism used to excise cut-and-paste transposons from DNA has been found to operate in the developing immune system of vertebrates, catalyzing the DNA rearrangements that produce antibody and T-cell receptor diversity. Known as V(D)J recombination, this process will be discussed in Chapter 24. Found only in vertebrates, V(D)J recombination is a relatively recent evolutionary novelty, but its mechanism was probably derived from the much more ancient cut-and-paste transposons.
Some DNA-only Transposons Move by Replicating Themselves
Although cut-and-paste transposition is common, especially in bacteria, there are other ways that DNA-only transposons can move. These involve replicating the transposon and moving the copy to a new position on the genome, leaving the original transposon intact and in its original position. There are several different ways this can occur and we discuss only one here, which is characteristic of a large class of DNA-only transposons known as helitrons. Found in all branches of life, these transposons are especially common in plants and animals where they can compose several percent of genomes. They carry a gene for an unusual type of transposase, one that functions as both a sequence-specific nuclease and as a helicase, thereby directing the movement of the transposon (Figure 5–60).
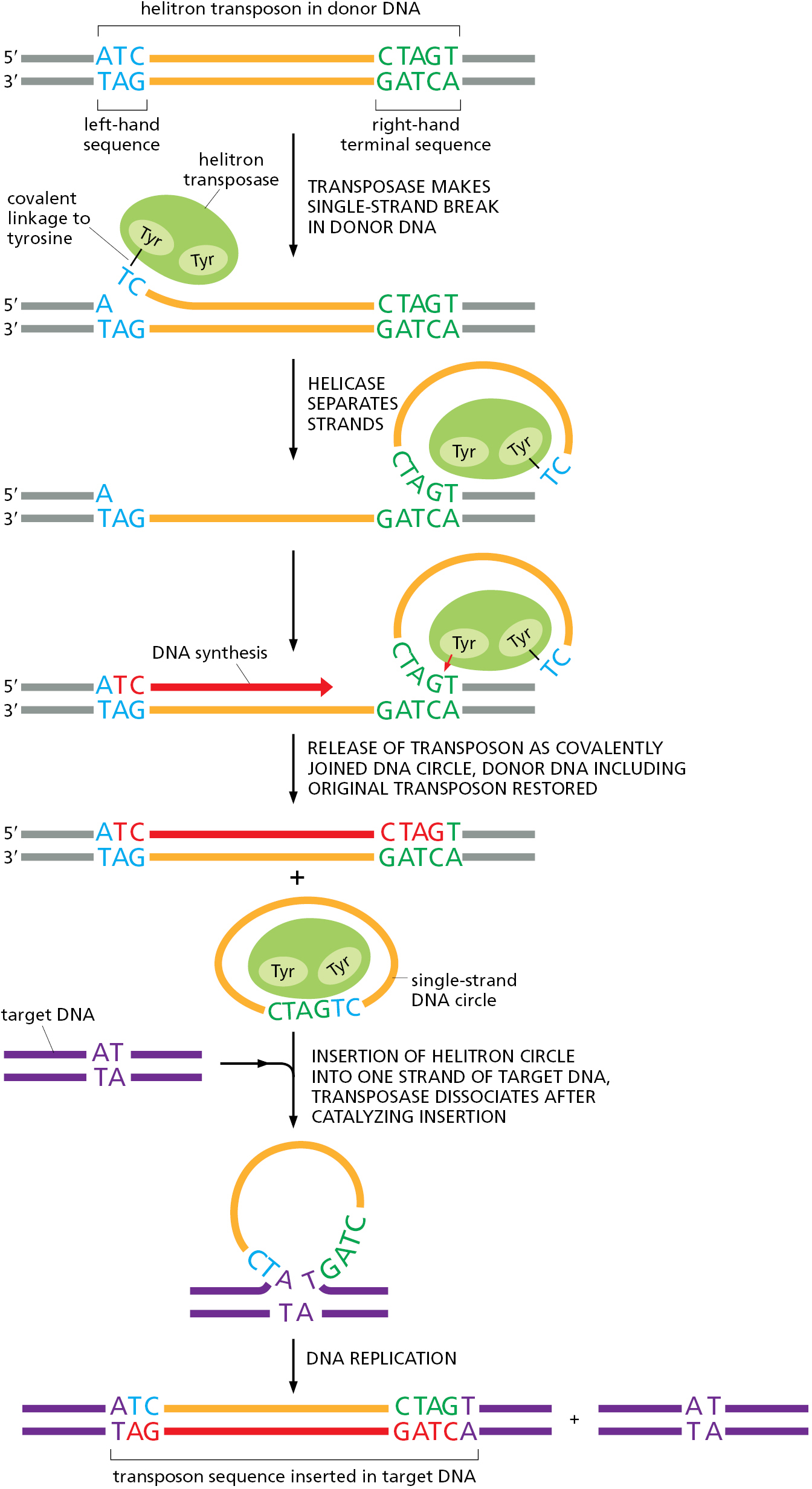
Because of the mechanism behind their movement, helitrons often transfer bits of the genome, along with themselves, to new positions. For this reason, they are thought to be especially important in reshuffling genomic information to produce variant organisms subject to natural selection over evolutionary time scales.
Some Viruses Use a Transposition Mechanism to Move Themselves into Host-Cell Chromosomes
Certain viruses are considered mobile genetic elements because they use transposition mechanisms to integrate their genomes into that of their host cell. However, unlike transposons, the nucleotide sequences that form these viruses encode proteins that package their genetic information into virus particles that can leave the original host cell to infect other cells. As discussed in Chapter 1, most viruses probably evolved from transposable elements through the capture of genes from their host cells. Although originally serving some other purpose in the cell, such captured genes, after a long process of mutation and selection, now code for the structural proteins of viruses, allowing them to escape the cell. Viruses are among the most numerous biological entities on Earth, and we discuss them in more detail in Chapter 23. The viruses that insert themselves into host chromosomes generally do so by employing one of the first two mechanisms listed in Table 5–4; namely, by behaving like DNA-only transposons or like retroviral-like retrotransposons. Indeed, much of our knowledge of these mechanisms has come from studies of particular viruses that employ them.
Transposition has a key role in the life cycle of many viruses. Most notable are the retroviruses, which include the human AIDS virus, HIV. Outside the cell, a retrovirus exists as a single-strand RNA genome packed into a protein shell, or capsid, along with a virus-encoded reverse transcriptase enzyme. During the infection process, the viral RNA enters a cell and is converted to a double-strand DNA molecule by the action of this crucial enzyme, which is able to polymerize DNA on either an RNA or a DNA template (Figure 5–61). The term retrovirus refers to the virus’s ability to reverse the usual flow of genetic information, which normally is from DNA to RNA (see Figure 1–4).
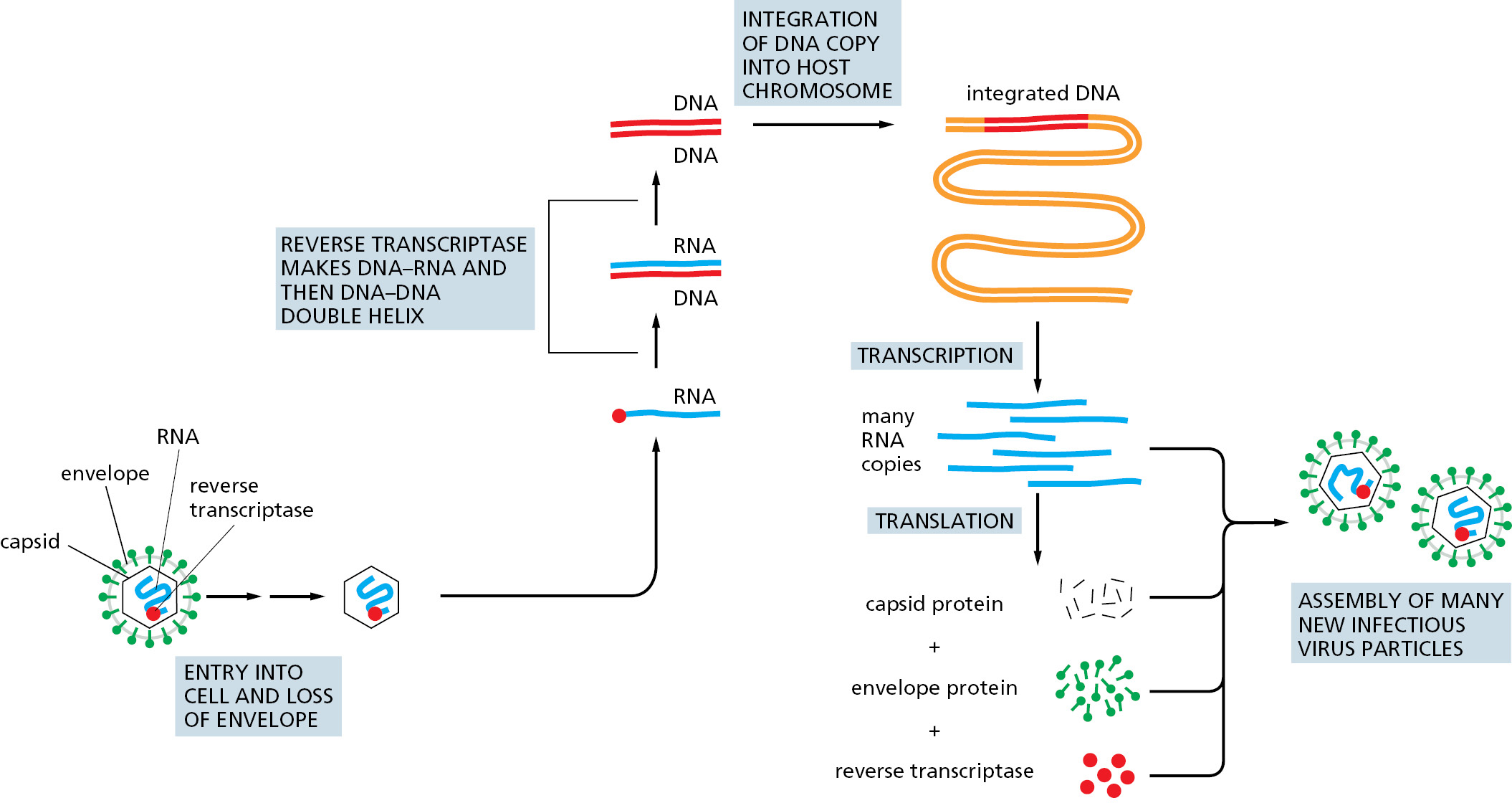
Once the reverse transcriptase has produced a double-strand DNA molecule, specific sequences near its two ends are recognized by a virus-encoded transposase called integrase. Integrase then inserts the viral DNA into the chromosome by a mechanism similar to that used by the cut-and-paste DNA-only transposons (see Figure 5–59).
Some RNA Viruses Replicate and Express Their Genomes Without Using DNA as an Intermediate
Retroviruses are not the only viruses that carry their genomes in the form of RNA. Other viruses also have single-strand RNA genomes, but, unlike retroviruses, many replicate and express their genomes without ever using DNA; that is, they are RNA-only viruses. For example, SARS-CoV-2, the coronavirus underlying the COVID-19 pandemic, replicates its single-strand RNA genome using a special, viral-encoded RNA-dependent RNA polymerase. Upon entering a cell, the viral genome is directly translated by ribosomes as though it were an mRNA molecule, producing many different viral-encoded proteins, including the RNA-dependent RNA polymerase. (We discuss mRNA and the process of translation in detail in Chapter 6.) The polymerase assembles with several other viral proteins and a few host proteins to form the complete replicase complex. This specialized replicase, which does not require a primer to begin synthesis, starts at the 3′ end of the viral genome and makes a complementary RNA copy of the entire genome (Figure 5–62). Using this complementary copy as a template, the replicase then synthesizes new genomes, which are then packaged with newly synthesized viral proteins into complete virus particles. The whole process of viral replication takes about 10 hours, and a single infected cell can produce as many as 1000 new virus particles, which can spread to other cells within the same host or move in aerosols to new hosts. Because coronaviruses do not use DNA, all steps of viral replication can take place outside the nucleus. In the case of SARS-CoV-2, viral replication occurs in the cytoplasm inside double-membrane compartments that are commandeered by the virus from the endoplasmic reticulum, an organelle described in detail in Chapter 12. These virus-induced compartments are believed to protect the virus from the cell’s many antiviral defenses (see pp. 1337–1338) during viral replication and assembly.
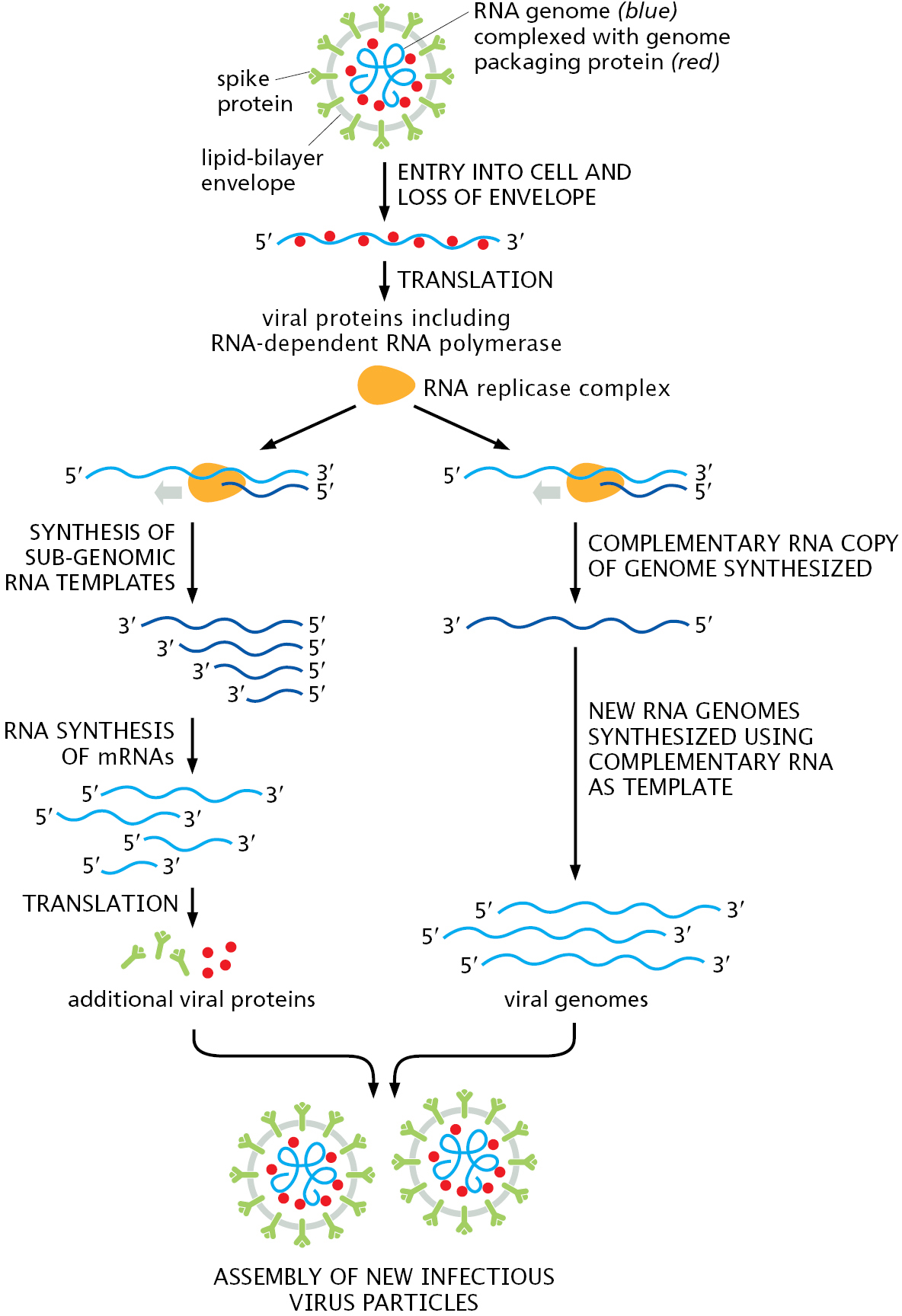
Among the first proteins made by coronaviruses are those that form the RNA-dependent RNA polymerase, which is responsible for producing new viral genomes by synthesizing RNA using RNA as a template. The replicase complex (which includes the polymerase and several other loosely associated proteins) first synthesizes complete noncoding copies of the viral genome. These complementary copies in turn serve as templates for the replicase complex to synthesize new genomes. The replicase also makes a series of shorter coding RNAs, which are needed to produce additional viral proteins including the spike. Once new viral genomes and proteins have been synthesized, new virus particles are assembled and exit the cell. Although only a few viral proteins are shown in the diagram, the virus codes for at least 27 different proteins; some of these organize the double-membrane structures in which the virus replicates, while others inhibit various immune system responses to the infection.
Several features of coronaviruses distinguish them from other RNA-only viruses such as those that cause influenza or polio. Perhaps the most unusual is the ability of coronavirus replicase complexes to proofread as they copy their RNA genomes. This proofreading occurs in much the same way that we saw for DNA polymerases earlier in the chapter: An incorrectly added nucleotide is excised by a 3′-to-5′ exonuclease carried in the replicase complex, giving the replicase another chance to add the correct nucleotide. This feature means that coronaviruses do not mutate as rapidly as most other RNA viruses, which lack proofreading ability. As discussed in Chapter 23, the relatively high mutation rate of influenza virus helps explain why new vaccines are needed every year, and this may not always be the case for coronaviruses.
The proofreading also has other important implications. As discussed earlier in this chapter, the lower the mutation rate, the greater the number of essential proteins that a genome can maintain. Proofreading allows coronaviruses to have a larger genome than is typical for RNA viruses; for example, compare the 30,000 nucleotides of the SARS-CoV-2 genome (coding for at least 27 proteins) to the 13,500 nucleotides of the influenza virus (coding for about 10 proteins). Proofreading also affects the development of antiviral drugs. Viral replicases are attractive targets for such drugs, in part because similar enzymes do not exist in uninfected host cells, reducing the chance of side effects. Drugs of this type (for example, remdesivir) are typically nucleoside triphosphate analogs that “fool” the RNA replicase into adding them to growing RNA chains. Once incorporated, the analogs—which have improper 3′ ends—poison further chain elongation (see Figure 8–42). Coronavirus proofreading can excise many of these analogs and thereby reduce their potency. A related strategy (exemplified by molnupiravir) employs nucleoside triphosphate analogs that are incorporated into RNA by the viral replicase, escape proofreading, but base pair incorrectly in the next round of replication, thereby introducing a lethal number of mutations.
Another striking feature of coronaviruses is the way in which they make many different proteins from a genome carried on a single RNA molecule. As described earlier, the viral genome, once it enters a cell, is treated like an mRNA molecule and translated into protein. We shall see in the next chapter, however, that most eukaryotic mRNAs can code for only a single protein. Coronavirus production of many different proteins from a single RNA genome requires a series of unusual steps, some of which appear unique to coronaviruses. We shall discuss the general topics of mRNA translation and its regulation in Chapters 6 and 7. But first, we return to our discussion of transposons, some of which closely resemble viruses in the way they move from place to place in their host genomes.
Retroviral-like Retrotransposons Resemble Retroviruses, but Cannot Move from Cell to Cell
A large family of transposons called retroviral-like retrotransposons (see Table 5–4) move themselves in and out of chromosomes by a mechanism similar to that used by retroviruses. These elements are present in organisms as diverse as yeasts, flies, and mammals; unlike viruses, they have no intrinsic ability to leave their resident cell but are passed along to all descendants of that cell through the normal processes of DNA replication and cell division. The first step in their transposition is the transcription of the entire transposon, producing an RNA copy of the element that is typically several thousand nucleotides long. This transcript, which is translated as a messenger RNA by the host cell, encodes a reverse transcriptase enzyme. This enzyme makes a double-strand DNA copy of the RNA molecule via an RNA–DNA hybrid intermediate, precisely mirroring the early stages of infection by a retrovirus (see Figure 5–61). Like a retrovirus, the linear, double-strand DNA molecule then integrates into a site on the chromosome using an integrase enzyme that is also encoded by the element. The structure and mechanisms of these integrases closely resemble those of the transposases of DNA-only transposons.
A Large Fraction of the Human Genome Is Composed of Nonretroviral Retrotransposons
A significant fraction of many vertebrate chromosomes is made up of repeated DNA sequences. In human chromosomes, these repeats are mostly mutated and truncated versions of nonretroviral retrotransposons, the third major type of transposon (see Table 5–4). Although most of these transposons in the human genome are immobile, a few retain the ability to move. Movements of the L1 element (sometimes referred to as a LINE, or long interspersed nuclear element) have been identified, some of which result in human disease; for example, a particular type of hemophilia results from an L1 insertion into the gene encoding the blood-clotting protein Factor VIII (see Figure 6–25).
Nonretroviral retrotransposons are found in many organisms and move via a distinct mechanism that requires a complex of an endonuclease and a reverse transcriptase. As illustrated in Figure 5–63, the RNA and reverse transcriptase have a much more direct role in the recombination event than they do in the retroviral-like retrotransposons described above.
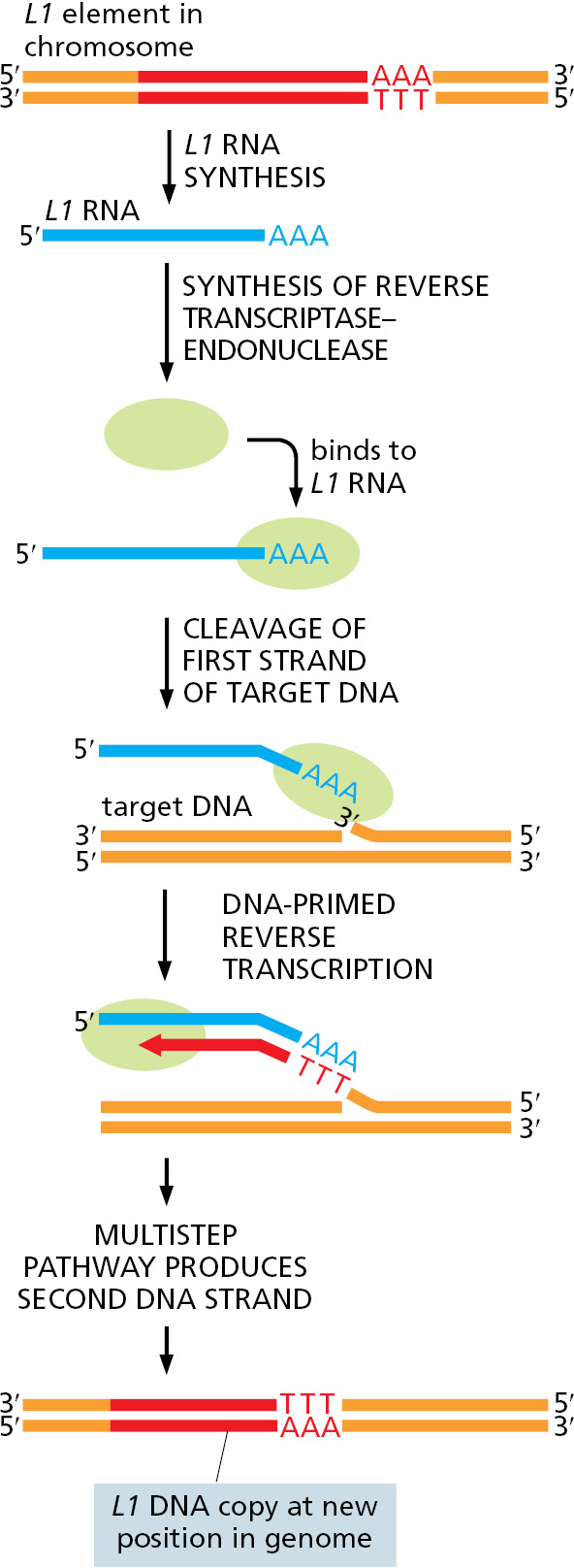
Inspection of the human genome sequence reveals that the bulk of nonretroviral retrotransposons—for example, the many copies of the Alu element, a member of the SINE (short interspersed nuclear element) family—do not carry their own endonuclease or reverse transcriptase genes. Nonetheless, they have successfully amplified themselves to become major constituents of our genome, presumably by pirating enzymes encoded by active L1 elements. Together the LINEs and SINEs make up more than 30% of the human genome (see Figure 4–62); there are 500,000 copies of the former and more than a million of the latter.
Different Transposable Elements Predominate in Different Organisms
We have described several types of transposable elements: (1) DNA-only transposons, the movement of which is based on DNA breaking and joining reactions; (2) retroviral-like retrotransposons, which also move via DNA breakage and joining, but where RNA has a key role as a template to generate the DNA recombination substrate; and (3) nonretroviral retrotransposons, in which an RNA copy of the element is central to the incorporation of the element into the target DNA, acting as a direct template for a DNA target–primed reverse transcription event.
Intriguingly, different types of transposons predominate in different organisms. For example, the vast majority of bacterial transposons are DNA-only types, with a few related to the nonretroviral retrotransposons also present. In yeasts, the main mobile elements are retroviral-like retrotransposons. In Drosophila, DNA-only, retroviral, and nonretroviral transposons are all found. Finally, the human genome contains all three types of transposon, but as discussed below, their evolutionary histories are strikingly different.
Genome Sequences Reveal the Approximate Times at Which Transposable Elements Have Moved
The nucleotide sequence of the human genome provides a rich fossil record of the activity of transposons over evolutionary time spans. By carefully comparing the nucleotide sequences of the approximately 3 million transposable element remnants in the human genome, it has been possible to broadly reconstruct the movements of transposons in our ancestors’ genomes over the past several hundred million years. For example, the cut-and-paste DNA-only transposons appear to have been very active well before the divergence of humans and Old World monkeys (25–35 million years ago), but because they gradually accumulated inactivating mutations, they have been dormant in the human lineage since that time. Likewise, although our genome is littered with relics of retroviral-like retrotransposons, none appear to be active today. Only a single family of retroviral-like retrotransposons is believed to have transposed in the human genome since the divergence of human and chimpanzee approximately 6 million years ago. The nonretroviral retrotransposons are also ancient, but in contrast to other types, some are still moving in our genome, as mentioned previously. For example, it is estimated that de novo movement of an Alu element occurs once in every 100–200 human births. This movement of nonretroviral retrotransposons is responsible for a small but significant fraction of new human mutations—perhaps two mutations out of every thousand.
The situation in mice is significantly different. Although the mouse and human genomes contain roughly the same density of the three types of transposons, both types of retrotransposons are still actively transposing in the mouse genome, being responsible for approximately 10% of new mutations.
Although we are only beginning to understand how the movements of transposons have shaped the genomes of present-day mammals, it has been proposed that bursts in transposition activity could have been responsible for critical speciation events during the radiation of the mammalian lineages from a common ancestor, a process that began approximately 170 million years ago. At present, we can only wonder how many of our uniquely human qualities have been derived from the past activity of the mobile genetic elements whose remnants are found scattered throughout our chromosomes.
Conservative Site-specific Recombination Can Reversibly Rearrange DNA
A different kind of recombination mechanism, known as conservative site-specific recombination, rearranges other types of mobile DNA elements. In this pathway, breakage and joining occur at two special sites, one on each participating DNA molecule, with the recombination event being carried out by a specialized enzyme that breaks and rejoins the two DNA double helices at these specific sequences. The same enzyme system that joins two DNA molecules can often take them apart again, precisely restoring the sequence of the two original DNA molecules (Figure 5–64A). Alternatively, with a different orientation of these two sequences in a chromosome, conservative site-specific recombination produces a DNA inversion (Figure 5–64B).
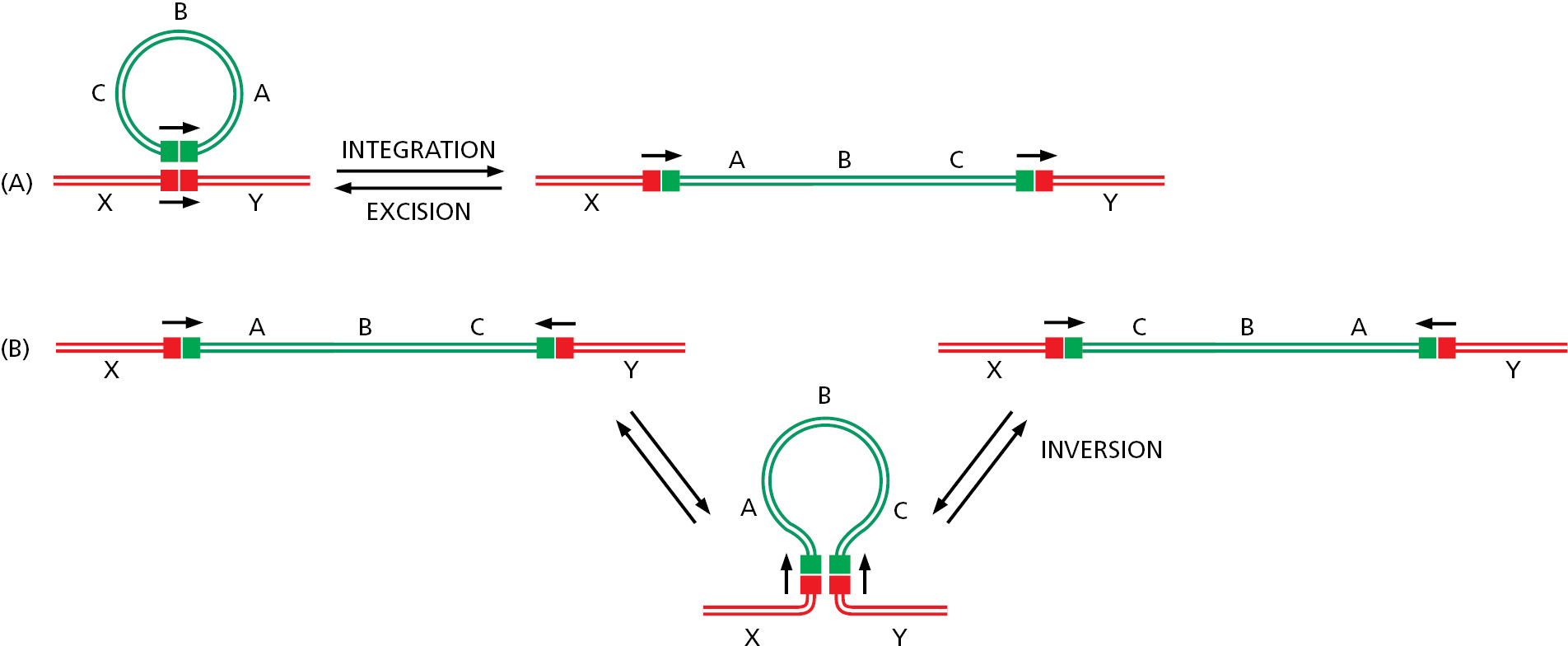
The conservative site-specific recombination pathway illustrated in Figure 5–64A is often used by bacterial DNA viruses to move their genomes in and out of the genomes of their host cells. When integrated into its host genome, the viral DNA is replicated along with the host DNA and is faithfully passed on to all descendant cells. If the host cell suffers damage (for example, by UV irradiation), the virus can reverse the site-specific recombination reaction, excise its genome, and package it into a virus particle. In this way, many viruses can replicate themselves passively as a component of the host genome, but can also “leave the sinking ship” by excising their genomes and packaging them in a protective coat until a new, healthy host cell is encountered.
Several features distinguish conservative site-specific recombination from transposition. First, conservative site-specific recombination requires specialized DNA sequences on both the donor and recipient DNA (hence the term “site-specific”). These sequences contain recognition sites for the particular recombinase that will catalyze the rearrangement. In contrast, transposition requires only that the transposon bears a specialized sequence; for most transposons, the recipient DNA can be of nearly any sequence. Second, the reaction mechanisms are fundamentally different. The recombinases that catalyze conservative site-specific recombination resemble topoisomerases in the sense that they form transient high-energy covalent bonds with the DNA and use this energy to complete all the DNA rearrangements without the need for new DNA synthesis (see Figure 5–22). Thus, all the phosphate bonds that are broken during a recombination event are restored upon its completion (hence the term “conservative”). Transposition, in contrast, typically leaves gaps in the DNA that must be repaired by DNA polymerases.
Conservative Site-specific Recombination Can Be Used to Turn Genes On or Off
Many bacteria use conservative site-specific recombination to control the expression of particular genes. A well-studied example occurs in Salmonella bacteria, an organism that is a major cause of food poisoning in humans. Known as phase variation, the switch in gene expression results from the occasional inversion of a specific 1000-nucleotide-pair piece of DNA, brought about by a conservative site-specific recombinase encoded in the Salmonella genome. This change alters the expression of the cell-surface protein flagellin, for which the bacterium has two different genes. The DNA inversion changes the orientation of a promoter (a DNA sequence that directs transcription of a gene) that is located within the inverted DNA segment. With the promoter in one orientation, the bacteria synthesize one type of flagellin; with the promoter in the other orientation, they synthesize the other type (Figure 5–65).
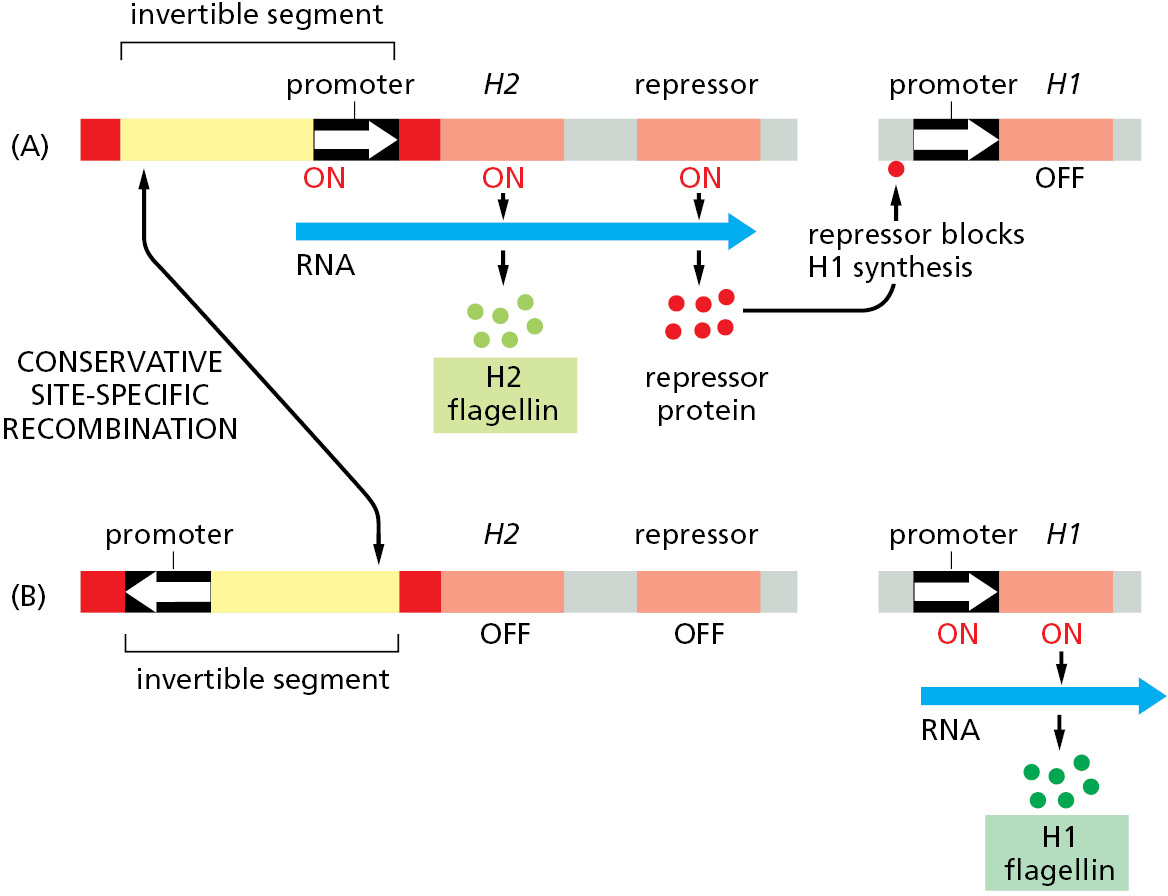
The recombination reaction is reversible, allowing bacterial populations to switch back and forth between the two types of flagellin. Inversions occur only rarely, and because such changes in the genome will be copied faithfully during all subsequent replication cycles, entire clones of bacteria will have one type of flagellin or the other.
Phase variation helps protect the bacterial population against the immune response of its vertebrate host. If the host makes antibodies against one type of flagellin, a few bacteria whose flagellin has been altered by gene inversion will still be able to survive and multiply.
Bacterial Conservative Site-specific Recombinases Have Become Powerful Tools for Cell and Developmental Biologists
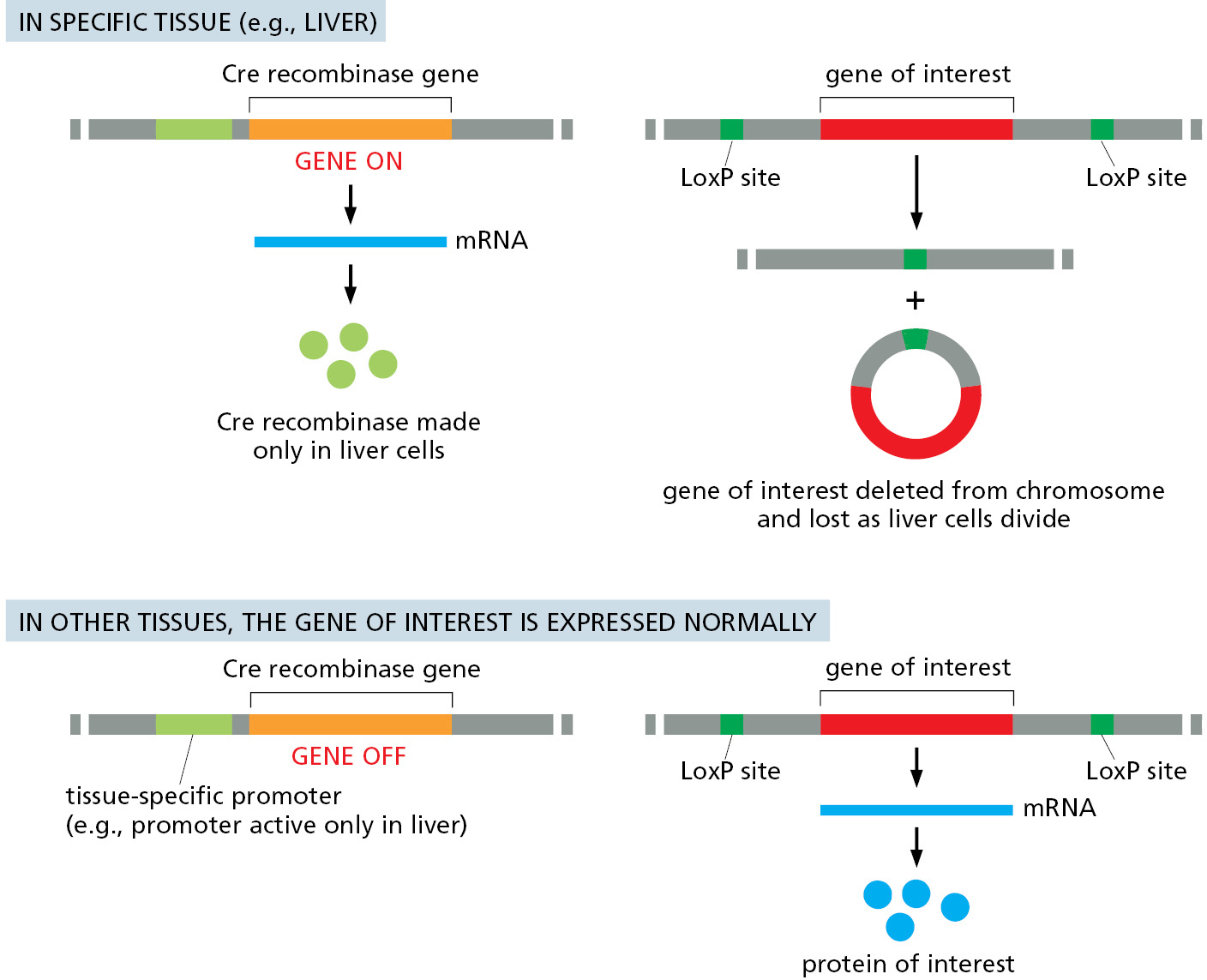
Like many of the mechanisms used by cells and viruses, site-specific recombination has been put to work by scientists to aid in the study of a wide variety of problems. To decipher the roles of specific genes in complex multicellular organisms, genetic engineering techniques are used to produce worms, flies, and mice carrying both a gene encoding a site-specific recombination enzyme and a carefully designed target DNA that includes a gene of interest flanked by DNA sites recognized by the recombination enzyme. At an appropriate time, the gene encoding the enzyme can be activated to rearrange the target DNA sequence. Such a rearrangement is widely used to delete a specific gene in a particular tissue of a multicellular organism (Figure 5–66). This strategy is particularly useful when the gene of interest plays a key role in the early development of many tissues, and a complete deletion of the gene from the germ line would cause death very early in development. The same strategy can also be used to artificially express any specific gene in a tissue of interest; here, the triggered deletion joins a strong transcriptional promoter to the gene of interest. With this tool one can in principle determine the influence of any protein in any desired tissue of an intact animal.
Summary
The genomes of nearly all organisms contain mobile genetic elements that can move from one position in the genome to another by either transposition or conservative site-specific recombination. In most cases, this movement is random and happens at a very low frequency. There are three classes of transposons: the DNA-only transposons, the retroviral-like retrotransposons, and the nonretroviral retrotransposons. The first two classes have close relatives among the viruses, including the human retrovirus that causes AIDS, HIV. Although mobile genetic elements can be viewed as parasites, many of the new arrangements of DNA sequences that their recombination events produce have been important for creating the genetic variation required for the evolution of cells and organisms.
Glossary
- transposition (transpositional recombination)
- Movement of a DNA sequence from one chromosomal site to another.
- conservative site-specific recombination
- A type of DNA recombination that takes place between short, specific sequences of DNA and occurs without the gain or loss of nucleotides. Unlike homologous recombination, it does not require an extensive region of homology between the two recombining DNA molecules.
- transposons
- Segment of DNA that can move from one chromosomal position to another by transposition.
- transposable element
- Segment of DNA that can move from one chromosomal position to another by transposition.
- DNA-only transposon
- Transposable element that exists as DNA throughout its life cycle. Many of these elements move by cut-and-paste transposition. See also transposon.
- retrovirus
- RNA-containing virus that replicates in a cell by first making an RNA–DNA intermediate and then a double-strand DNA molecule that becomes integrated into the cell’s DNA.
- reverse transcriptase
- Enzyme first discovered in retroviruses that makes a double-strand DNA copy from a single-strand RNA template molecule.
- phase variation
- The random switching of phenotype of an infectious agent that is caused by changes in expression of proteins at frequencies much higher than mutation rates.


