A. Streptomyces venezuelae
eResearch Activity 3
How Do a Hundred Cells Divide at Once?
Dividing the cell is a complicated business, even for a relatively simple bacterium such as Escherichia coli. Forming two functional daughter cells requires intricate coordination of the DNA replication and cell growth, culminating in septation at the Z-ring (see Figures 3.23 and 3.25). Now suppose you have to form a hundred cells at once. How would that ever work?
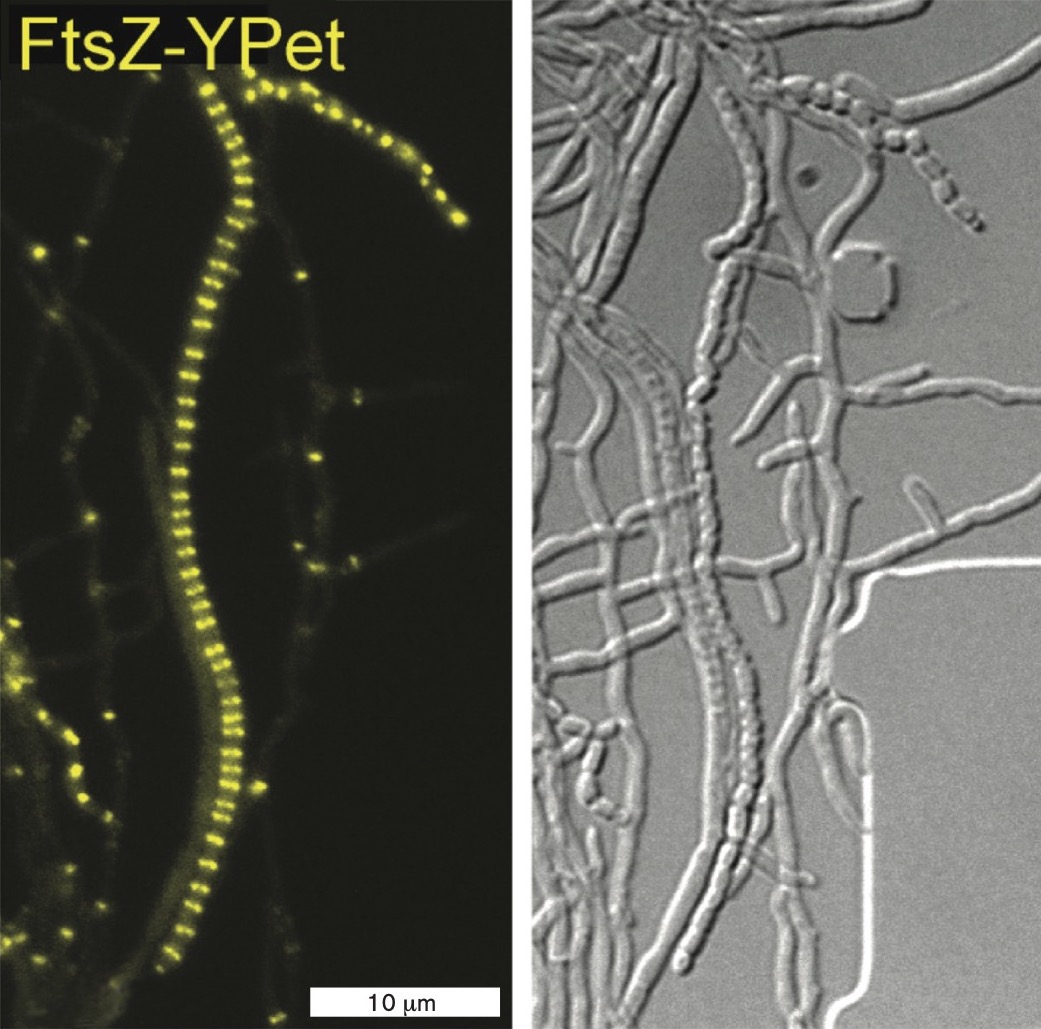
The actinomycete Streptomyces venezuelae grows as a filament (or hypha), copying its DNA many times and extending without cell division. In a sporogenic (spore-forming) hypha, a molecular regulator promotes simultaneous septation at dozens of Z-rings. When FtsZ is labeled with a fluorophore, the fluorescence generates a dramatic ladder-like pattern called a “Z-ladder” (Fig. ERA 3.1A  ). The Z-ladder enables a filament to divide and develop into numerous spores, which are carried off on air currents to colonize a new location. Streptomyces species are important to understand because they give us many antibiotics, such as streptomycin for tuberculosis and tetracycline for acne and respiratory bacterial infections.
). The Z-ladder enables a filament to divide and develop into numerous spores, which are carried off on air currents to colonize a new location. Streptomyces species are important to understand because they give us many antibiotics, such as streptomycin for tuberculosis and tetracycline for acne and respiratory bacterial infections.
A. S. venezuelae
More information
A fluorescent micrograph of a sporogenic hyphae of S. venezuelae. The hyphae is shown as a 45 micrometer line and a 15 micrometer line composed of Z rings. Both lines are long and fairly straight.
B. S. venezuelae ΔsepH
More information
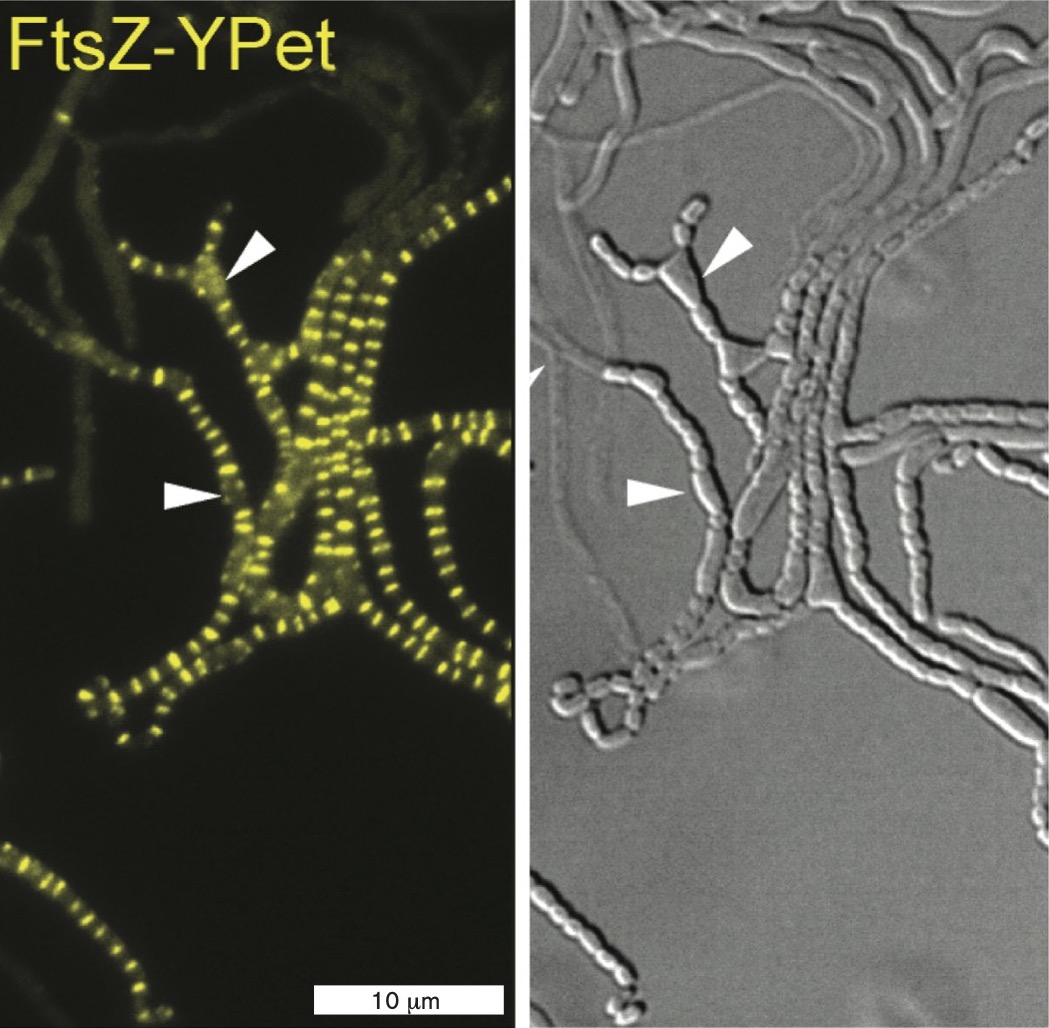
A fluorescent micrograph of a sporogenic hyphae of a mutated S. venezuelae. The strain has had the s e p H gene deleted. Instead of long, independent hyphaes, this cell line has a cluster of line fragments that are approximately 20 micrometers long.
 ; Panel B:
; Panel B:  )F. RAMOS-LEÓN ET AL. 2021. ELIFE. 10:E63387F. RAMOS-LEÓN ET AL. 2021. ELIFE. 10:E63387
)F. RAMOS-LEÓN ET AL. 2021. ELIFE. 10:E63387F. RAMOS-LEÓN ET AL. 2021. ELIFE. 10:E63387B. Streptomyces venezuelae ΔsepH
How do Streptomyces hyphae manage to coordinate a hundred cell divisions? Susan Schlimpert, at the John Innes Centre in Norwich, England, investigates the mechanisms of cell division in Streptomyces venezuelae, a model actinomycete (Fig. ERA 3.2A). A key cell division regulator was revealed by her postdoctoral fellow Félix Ramos-León and her research assistant Matthew Bush (Fig. ERA 3.2B).
More information
A photo of Susan Schlimpert. She has short blonde hair and a dark red shirt on. She appears in a lab setting.
A photo of Felix Ramos Leon and Matthew Bush. Felix has dark hair, a goatee and a dark orange shirt. Matthew has dark hair, glasses, a goatee and a dark plaid shirt.
Bush identified sepH as a gene of unknown function in the Streptomyces genome and showed that its expression is controlled by a major sporulation regulator. First, Ramos-León and colleagues investigated the SepH gene product by constructing a gene fusion to the gene encoding the fluorescent protein YPet. (Gene fusions for fluorescent proteins are discussed in Chapter 2.) The resulting fusion gene, sepH-ypet, encodes a protein that functions as SepH but also fluoresces as YPet by absorption of green light with emission of yellow light. In microscopy of S. venezuelae carrying the sepH-ypet gene, we see yellow fluorescence at the position of Z-rings. Thus, SepH has an appropriate location for a protein that assists FtsZ function.
Next, the researchers observed a strain of S. venezuelae that expresses FtsZ fused to YPet, so that the ladder of Z-rings fluoresces. The FtsZ-YPet fluorescence was observed in two different strains, either expressing sepH (Fig. ERA 3.1A) or deleted for sepH (ΔsepH) (Fig. ERA 3.1B  ). For each experiment, a movie was made showing the coordinated development of the Z-ladder and sporulation. In the ΔsepH strain (a strain lacking sepH), sporulation becomes disorganized, causing cells to bulge into distorted forms and even to form filament branches. While some sporulation occurs, the process is greatly disrupted. Thus sepH was confirmed as a gene whose protein product organizes septation and sporulation in S. venezuelae.
). For each experiment, a movie was made showing the coordinated development of the Z-ladder and sporulation. In the ΔsepH strain (a strain lacking sepH), sporulation becomes disorganized, causing cells to bulge into distorted forms and even to form filament branches. While some sporulation occurs, the process is greatly disrupted. Thus sepH was confirmed as a gene whose protein product organizes septation and sporulation in S. venezuelae.
Ramos-León further revealed that SepH stimulates the GTPase activity of FtsZ, a common mechanism of molecular regulation. A SepH homolog was found in the genus Mycobacterium, a group of Actinobacteria that includes the famous pathogen Mycobacterium tuberculosis. In pathogenic mycobacteria, SepH assists cell division without sporulation. Thus, discovery of this cell division protein advances our understanding of both an important drug producer and a major pathogen whose mechanisms may lead to novel antibiotic therapies.
Further Exploration
What experiments might you perform to show whether SepH interacts directly with the Z-ladder by binding to FtsZ? What other kinds of proteins might be needed?
Ramos-León, Félix, Matthew J. Bush, Joseph W. Sallmen, Govind Chandra, Jake Richardson, et al. 2021. A conserved cell division protein directly regulates FtsZ dynamics in filamentous and unicellular actinobacteria. Elife 10:e63387.
Glossary
- Figure 3.23
-
More information
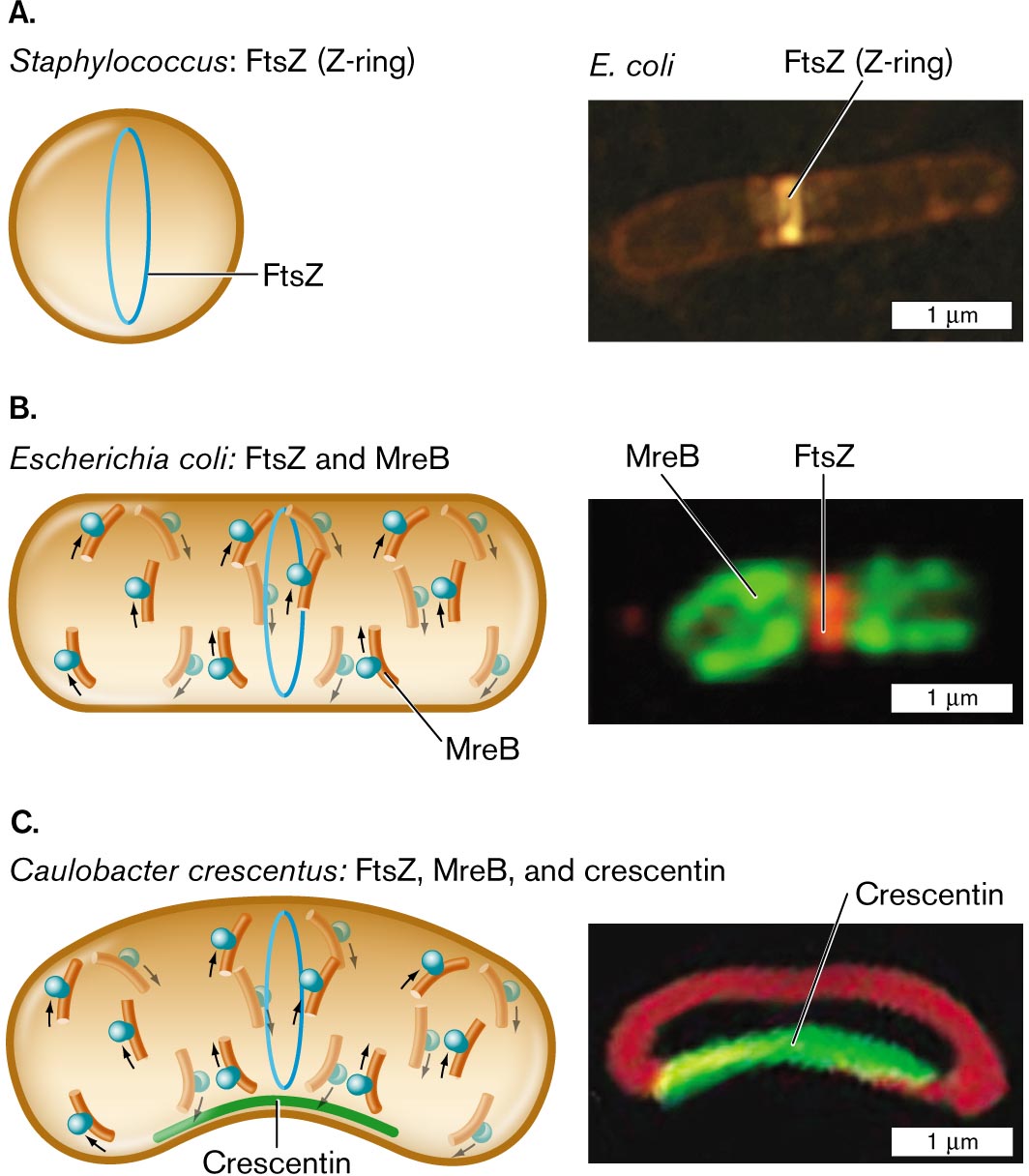
Three illustrations and accompanying fluorescence micrographs describing shape determining proteins in different bacteria.
An illustration and an accompanying fluorescence micrograph of F t s Z in Staphylococcus. The cell is circular shaped, and inside the cell is a vertical oval ring labeled F t s Z. To the right is a microscopic image of F t s Z, or Z-ring, fluorescing in an E. coli bacterium. E. coli is a rod shaped cell of about 3 micrometers in length and 0.5 micrometer in width. F t s Z is identified as a bright band near the center of the cell.
An illustration and an accompanying fluorescence micrograph of F t s Z and M r e B in Escherichia coli. The cell is a long horizontal rod shape. The F t s Z ring is in the center, and overlapping and to the left and right are M r e B rings. To the right shows a microscopic fluorescent image of F t s Z bonded in the center of a cell and M r e B at the edges. The unit is about 2 micrometers long and 1 micrometer wide.
An illustration and an accompanying fluorescence micrograph of F t s Z, M r e B, and crescentin in Caulobacter crescentus. The cell is a long horizontal rod shape with a central bend. The bottom part of the cell where it curves is labeled Crescentin. F t s Z ring is located in the center of the cell and overlapping, and to the left and right are M r e B rings. To the right is a microscopic fluorescent image of a rod shaped cell with a central bend. The bend is fluorescent and labeled Crescentin. The cell is about 2 micrometers long and 1 micrometer wide.
FIGURE 3.23 ■ Shape-determining proteins. A. Cell diameter is maintained by FtsZ polymerization to form the Z-ring. B. Elongation of a rod-shaped cell requires MreB proteins. MreB polymerizes around an E. coli cell (MreB-YFP fluorescence) along with a Z-ring of FtsZ (fluorescent anti-FtsZ antibody). C. Crescent-shaped cells possess a third shape-determining protein, CreS (crescentin), which polymerizes along the inner curve of the crescent. Crescentin protein fused to green fluorescent protein (CreS-GFP) localizes to the inner curve of Caulobacter crescentus. Membrane-specific stain FM4-64 (red fluorescence) localizes to the membrane around the cell. Q. SUN AND W. MARGOLIN. 1998. J. BACTERIOL. 180:2050 PURVA VATS AND LAWRENCE ROTHFIELD. 2007. PNAS 104:17795 NORA AUSMEES ET AL. 2003. CELL 115:705 - Figure 3.25
-
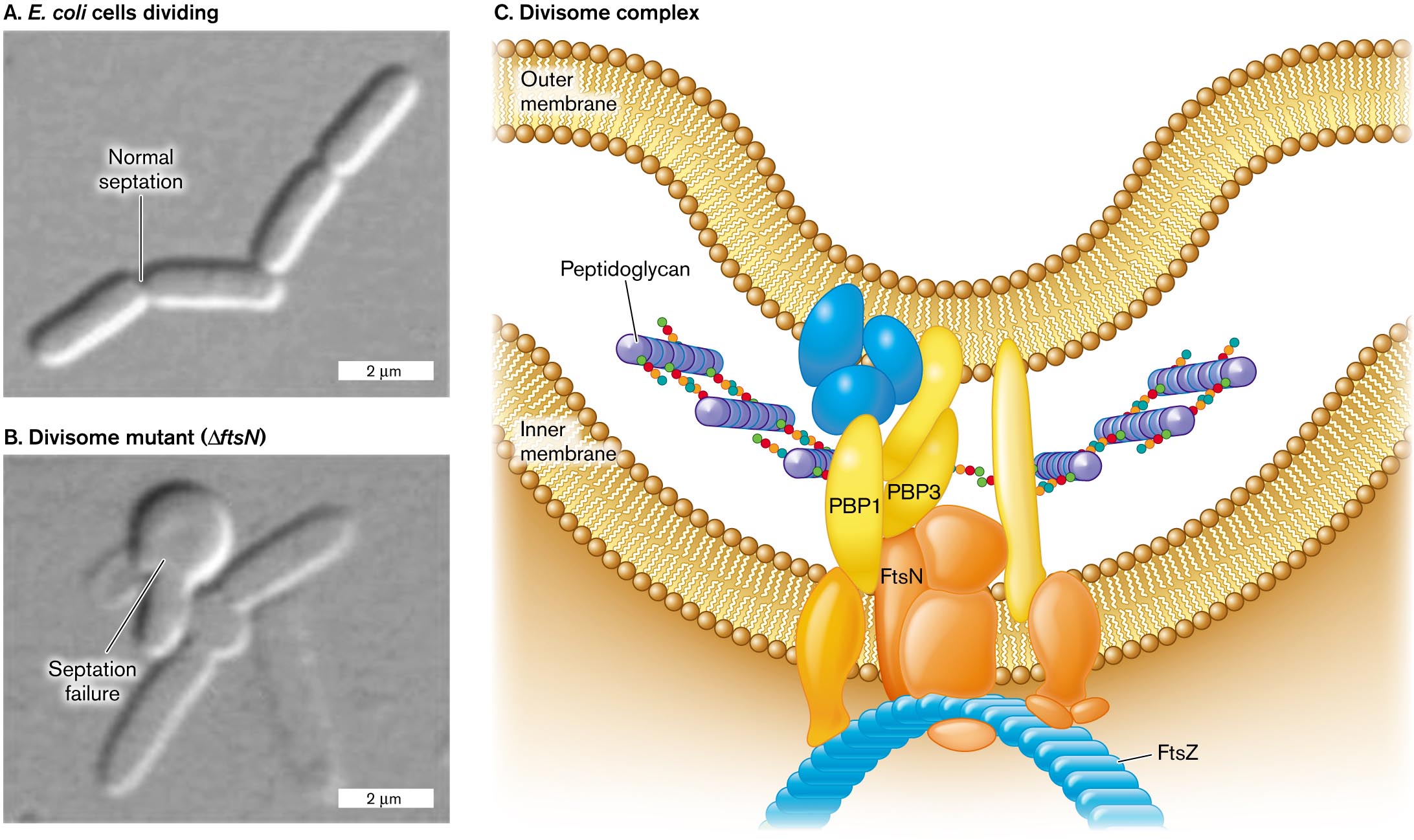
More information
Two micrographs of cell division and an illustration of the divisome complex.
A micrograph of E. coli cells dividing with normal septation. Four rod shaped cells in a chain are visible, each about 3 micrometers long and 0.5 micrometer wide. A split between the first two cells is labeled normal septation.
A micrograph of E. coli septation failure in a divisome mutant. A chain of a few rod shaped cells is visible, with a twisted clump of cells at the center of the chain. The clump is labeled septation failure. The divisome mutant is labeled delta f t s N.
An illustration of the structure of the divisome complex. The complex is located between the inner and outer membrane. Peptidoglycan synthesis is visible, occurring between P B P 1 and P B P 3. The divisome complex consists of several irregularly shaped proteins. F t s N is identified within the complex, and at the base of the complex where the cell shape can be seen curving, F t s Z is identified.
FIGURE 3.25 ■ Divisome enables cell division. A. E. coli cells divide normally. B. Cells that lack FtsN (divisome component) balloon out at the septum. C. The divisome complex coordinates the extension of all envelope layers while the septum constricts. Source: Part C modified from C. Typas et al. 2012. Nat. Rev. Microbiol. 10:123, fig. 2.
B. LIU ET AL. 2015. MOL. MICROBIOL. 95:945B. LIU ET AL. 2015. MOL. MICROBIOL. 95:945